Article Text
Abstract
Clonal haematopoiesis (CH) is defined by the presence of acquired mutations and/or cytogenetic abnormalities in haematopoietic cells. By definition, these premalignant clones do not meet criteria for haematopoietic neoplasms listed in the Revised Fourth Edition of the WHO classification. CH is fairly common in elderly individuals and is associated with higher risks for haematological cancers, in particular myelodysplastic syndrome and acute myeloid leukaemia (AML), as well as cardiovascular events. Similar small clones have also been detected during follow-up in patients with AML in morphological remission, in individuals with aplastic anaemia, and in pre-chemotherapy blood samples from patients with other types of cancers. In each of these contexts, the presence of mutations carries different clinical implications, and sometimes demonstrates unique genetic profiles. Emerging research suggests that the number and identity of mutations, the size of the mutant clones and various other factors, including age, immune status and history of exogenous drugs/toxins, are important for disease biology and progression. This review focuses specifically on the subset of CH with gene mutations detected by sequencing, and includes discussions of nomenclature and molecular technologies that detect and quantify gene mutations.
- molecular genetics
- leukocytes
- haematopathology
Statistics from Altmetric.com
Introduction
The concept of clonal haematopoiesis (CH) relates to the natural history of myeloid neoplasms, before the development of overt morphological and clinical features. CH is defined by the presence of acquired mutations and/or cytogenetic alterations in haematopoietic cells, in the absence of significant morphological abnormalities and often in the context of normal peripheral blood counts. By definition, these clones do not meet criteria for haematopoietic neoplasms in the current WHO classification.1 CH is fairly common in elderly individuals and is associated with elevated risks for haematopoietic malignancies,2–4 in particular myelodysplastic syndrome (MDS) and acute myeloid leukaemia (AML). As high-throughput sequencing becomes increasingly widespread, a working understanding of CH in different contexts can be helpful to pathologists who are increasingly likely to encounter these in practice. This short review focuses on the subset of CH with gene mutations detected by sequencing, which is often referred to as CH of indeterminate potential (CHIP) or age-related CH (ARCH) in the literature; we discuss this and other nomenclatures used to describe the phenomenon of CH, the technologies used to detect and quantify mutations, as well as more recent research. Readers interested in CH with cytogenetic abnormalities, which is not the focus of this manuscript, may consider additional recent reviews.5 6
The association between clonal leukocytes and ageing was first suggested by X-inactivation studies in women. During female embryonic development, the maternal or paternal X chromosomes are inactivated at random, which in principle, results in a roughly 50:50 pattern of X-inactivation in the cells of adults. What researchers found, however, was that these ratios in the blood leukocytes from some women were skewed and that the proportion of people with skewing increased with age.7 Several non-mutually exclusive explanations had been put forth to explain this age-associated skewing (AAS). One hypothesis pointed to decreasing numbers of haematopoietic stem cells in older people, suggesting that the skewing is caused by small numbers of precursor cells with genetic drift. Another hypothesis proposed that haematopoietic stem cells in some older individuals acquire gene mutations that promote proliferation and/or survival, causing mutated clones to preferentially expand over their wild-type counterparts. Support for the clonality hypothesis came in 2012, when Busque et al 8 found somatic mutations in TET2, a gene important for the pathogenesis of myeloid neoplasms, in the blood samples from a subset of elderly women with AAS.
Following this, in 2014, three research groups separately published analyses of exome sequencing performed on blood samples from large cohorts of individuals without known haematological malignancies.2–4 These investigators found recurrent mutations in genes known to be associated with myeloid neoplasms, most frequently DNMT3A, TET2 and ASXL1, in small but not insignificant percentages of people. As with AAS, the percentage of people with peripheral blood mutations increased with age: CH was found to be uncommon in those younger than 50 years,3 but was present in 10% or more of those older than 70 years.2 In contrast to the mutational profiles of MDS and AML,9–13 CH demonstrated the following: (1) fewer total numbers of mutations, (2) a more restricted set of involved genes (ie, mainly a few epigenetic regulators) and (3) lower mutant variant allele fractions (VAFs, usually <10%), consistent with smaller and genetically simpler clones. There was about a 10-fold elevated risk for myeloid malignancies among those with CH in comparison to those without CH2 3; however, the overall rate of progression to overt leukaemia was relatively low, estimated at 0.5%–1.0% per person per year, similar to the progression rates of monoclonal gammopathy of undetermined significance (MGUS)14 and monoclonal B-cell lymphocytosis (MBL).15 Individuals with CH also experienced shorter survival during follow-up.2 3 Interestingly, this finding was not explained by deaths from haematopoietic cancers, as there were relatively few such events in the cohorts. Relevant to this observation, Jaiswal et al 2 showed that CH was also associated with an increased risk for cardiovascular events. More recent research argue that this association is likely causal16 17 (see below, for further discussion of the relationship between CH and cardiovascular disease [CVD]).
Nomenclature for CH
Before proceeding further, it is important to point out there have been a variety of terms used to describe CH in the literature. In comparison to entities in the WHO classification, this nomenclature is not as well established, and different terms are often used interchangeably. Two frequently encountered acronyms are CHIP and ARCH. Table 1 lists definitions for these and other related terms.
Common terms encountered in the literature
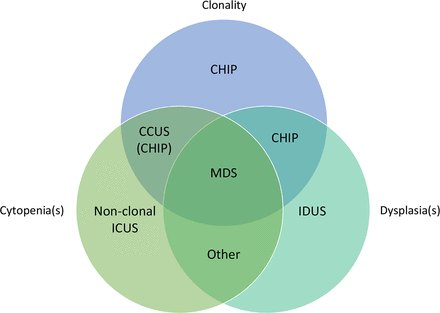
According to one reference,18 CHIP must fulfil two requirements: (1) the presence of acquired mutation(s) in genes associated with haematopoietic neoplasms, provisionally at a VAF >2% and (2) overall findings not meeting criteria for any WHO-defined entity, including MGUS and MBL, as well as other diseases that can have small haematopoietic clones, such as paroxysmal nocturnal haemoglobinuria. As defined, CHIP typically refers to elderly individuals with mutations who do not have cytopenias. The subset of cases with both mutations and cytopenia(s) may be more specifically designated as ‘clonal cytopenia of undetermined significance’ (CCUS) (figure 1), which are associated with genetically more complex clones and higher risk for progression to myeloid malignancies compared to cases of CHIP without cytopenia.19–21 ‘Idiopathic cytopenia of undetermined significance’ (ICUS, or more precisely non-clonal ICUS) is defined by unexplained cytopenia(s) in the absence of evidence of clonality or defining features of a WHO entity (figure 1), and this appears to have a lower risk for malignant progression than CCUS.20
Venn diagram illustrating the relationship between clonal haematopoiesis and related entities with respect to morphological dysplasia, cytopenia(s) and clonality. Please note that the Venn diagram is not to scale; for example, CHIP is much more common than MDS. For this figure, cytopenia and dysplasia are as defined in table 1; clonality refers to the presence of mutations and/or cytogenetic abnormalities, whether or not these are detected by testing. The diagnostic criteria for specific MDS entities are detailed in the WHO classification.1 In one commonly cited reference, CCUS is considered to be a form of CHIP18; however, others have proposed that CCUS and CHIP should be distinguished.27 The ‘other’ category includes non-clonal causes of cytopenia and abnormal morphology (eg, copper deficiency). CCUS, clonal cytopenia of undetermined significance; CHIP, clonal haematopoiesis of indeterminate potential; ICUS, idiopathic cytopenia of undetermined significance; IDUS, idiopathic dysplasia of undetermined significance; MDS, myelodysplastic syndrome.
Another commonly encountered four-letter acronym, ARCH, largely overlaps with CHIP, and emphasises the relationship between CH and ageing.5 However, it is important to note that CH is not limited to older individuals and can occur in other contexts; recent studies have identified small clones in patients with aplastic anaemia (AA)22 and those being treated with chemotherapy.23–26 In these settings, the presence of CH appears to be shaped by selective pressures (autoimmune and cytotoxic, respectively), and can occur in younger people.
Figure 1 illustrates the nomenclature for CH in relations to morphological abnormalities(s), cytopenia(s) and clonality. Notably, CH is not an official entity in the Revised Fourth Edition of the WHO classification1; it is distinguished from myeloid neoplasms by the absence of significant morphological findings (dysplasia or increased blasts) and/or cytopenia(s). Since the spectrum of mutations in CH overlaps with MDS and other myeloid neoplasms, gene mutations cannot be used at this time to reliably distinguish CH from MDS or other WHO entities. One consequence of this is that most gene mutations identified by high-throughput sequencing are not used as diagnostic biomarkers in the current WHO classification,1 although these mutations may contribute relevant prognostic and predictive information. Nevertheless, recent research and guidelines raise the possibility that the pattern of gene mutations can be helpful in the diagnosis of MDS cases with borderline morphologic features.20 27 It is possible that our existing ideas and nomenclature regarding CH and myeloid neoplasms will evolve as new research and frameworks emerge.
Sequencing technologies used to detect CH
Different molecular approaches used to characterise CH are summarised in table 2. In this section, we focus on platforms used to detect point mutations and small insertions, duplications, and deletions. Although larger genomic alterations (eg, a deletion demonstrated by conventional karyotype) are also important for the biology of myeloid neoplasms,28 29 cytogenetic testing will not be covered in this review.
Molecular technologies used for the assessment of CH: a comparison of approaches
The mutational profiles of CH were defined by comprehensive whole exome sequencing (WES, ie, the sequencing of all coding regions in the human genome).2–4 These exploratory studies provided proof of clonality and helped to establish the reference mutational landscape. Although comprehensive, the disadvantages of WES include high costs per sample and complexity; in addition, WES can be insensitive and may miss low-level driver mutations (eg,<2% VAF) that are clinically important (see below).
In clinical laboratories at the present time, the most common approach available for the evaluation of CH and myeloid neoplasms is targeted next-generation sequencing (NGS). These NGS panels focus on genes and exons recurrently mutated in haematopoietic neoplasms; since the regions sequenced are much smaller than the entire exome, greater sequencing depths at targeted loci can be achieved, potentially improving analytical sensitivity and lowering the limit of detection.
To further improve sensitivity for low-level mutations (eg, for the detection of minimal residual disease (MRD) and other settings) some sequencing technologies additionally incorporate molecular barcodes or similar tag sequences that uniquely label individual DNA templates.23 25 26 30–34 With these unique labels, investigators were better equipped to discriminate true low-level mutations from low-level technical artefacts that arise during the course of sequencing. NGS with molecular barcodes or molecular inversion probes (MIPs, which have analogous tag sequences) can detect and precisely quantify small clones with VAFs<0.1%, a level of sensitivity comparable to quantitative and droplet digital PCRs34–37 (table 2). In comparison to PCR-based methods, NGS with molecular barcodes or MIPs are more comprehensive, and are not limited to the assessment small numbers of ‘hotspot’ mutations, which is a major disadvantage of PCR (table 2).
Not surprisingly, the degree of analytical sensitivity can impact results. For example, several studies31 34 using more sensitive techniques have shown that the incidences of CH may be much higher than that detected in earlier studies using WES. Recent research has also linked low-level mutations (<0.1% VAFs) in pre-treatment blood samples from patients with non-myeloid cancers with the subsequent development of therapy-related myeloid neoplasms (t-MNs) following chemotherapy with or without autologous stem cell transplantation23 25 26; notably, many of these low-level mutations are below the limit of detection for WES. We will return to the topic of selecting an appropriate method to detect mutations in the section ‘Should we test for CH at this time?’
Progression from CHIP to MDS to AML
Sequencing studies of CHIP2–4 and myeloid neoplasms, including MDS9–11 and AML,12 13 have identified recurrent mutations in genes involved in diverse biological processes, including DNA methylation, histone modification, RNA splicing, tumour suppression (eg, via TP53), cell signalling, the cohesin complex and transcriptional regulation.
A comparison of the mutational profiles of CHIP and myeloid neoplasms at different stages of evolution reveal interesting patterns. For example, mutations in CHIP tend to occur in genes coding for specific epigenetic regulators.2–4 Epigenetics refers to modifications to DNA and histones (proteins that help ‘package’ DNA) that impact gene expression. DNMT3A (coding for DNA methyltransferase 3A) and TET2 (which affects DNA demethylation) are involved in DNA methylation; mutations in these genes change global methylation patterns, and alter the expression of many other genes, including those important for myeloid function and differentiation.28 ASXL1 regulates histone modifications, which also impacts gene transcription.28 CHIP in older individuals without cytopenias most often involve a single mutation in one of these genes, at a low VAF (usually <10%).2–4
By comparison, haematopoietic clones detected in the blood of patients with cytopenia(s) (ie, CCUS) often affect multiple genes at higher VAFs (often >10%).19–21 The probability of finding mutations in patients with cytopenias is also significantly higher than that in the general elderly population. The spectrum of mutations and VAFs in CCUS more closely overlaps with MDS, and includes mutations in spliceosome genes, with the exception of SF3B1.19–21 Depending on the specific pattern of mutations, CCUS may demonstrate a much higher risk for progression20 to overt myeloid malignancies than CHIP without cytopenia.
In comparison to CCUS, MDS has more frequent mutations in SF3B1,9–11 which are closely associated with the presence of ring sideroblasts in all WHO myeloid neoplasm subtypes. In MDS and myelodysplastic/myeloproliferative neoplasms, the presence of additional cooperating mutations in genes involved in cell signalling (including NRAS and FLT3), transcription (including RUNX1 and NPM1), as well as in IDH1 and IDH2, is associated with increased blasts and/or rapid progression to AML.28
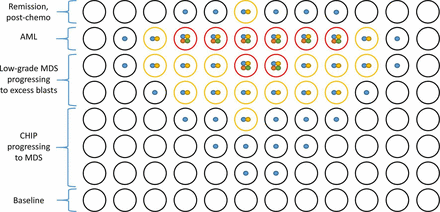
Accordingly, one plausible model is that mutations involving DNMT3A, TET2 and ASXL1 (ie, those common in CHIP) are early events in the pathogenesis of myeloid neoplasms, while the acquisition of mutations in genes coding for transcription factors or those involved in signalling are later events, heralding impending transformation to acute leukaemia (figure 2). With respect to this hypothesis, some have recently proposed that the subset of CH with gene mutations that show greater specificity for true myeloid neoplasms could be referred to as ‘CH of oncogenic potential,’38 which is to be distinguished from typical CHIP in the elderly. This pattern of early and late events raises the possibility that CH can be genetically risk stratified. Indeed, initial publications had shown that individuals with larger and more complex CHIP clones have a higher risk of subsequent myeloid neoplasms, although there was insufficient statistical power at that time to link mutations in specific genes to risk for AML.39
Morphology, mutations and the evolution of a myeloid neoplasm in a hypothetical individual. Each row is a snapshot of the clonal architecture at a specific time. Haematopoietic cells at each time point are represented by circles. The dots within the circles represent mutations. Black circles without dots are normal haematopoietic cells. The blue dots are early events in the pathogenesis of myeloid neoplasms (eg, a mutation in DNMT3A, TET2 or ASXL1). At this stage, there is no overt dysplasia in mutated cells (black). As the clone expands, it may acquire additional mutations (yellow/gold dot, eg, a mutation in SF3B1) associated with morphological abnormalities (yellow cells), ineffective haematopoiesis and cytopenias, enabling pathologists to formally diagnose MDS. The acquisition of late genetic events (green and red dots, eg, including a mutation involving FLT3) is associated with rapid progression of disease and increased blasts (red cells), leading to AML. Following chemotherapy for AML, residual mutant clones may be detected by sequencing. Please note that only a small percentage of individuals with CHIP will develop a WHO-defined myeloid neoplasm. AML, acute myeloid leukaemia; CHIP, clonal haematopoiesis of indeterminate potential; MDS, myelodysplastic syndrome.
More recently in 2018, two groups40 41 explored the association between antecedent mutations in CH and the risk for AML in greater detail. Banked blood samples from sizeable cohorts several years before the development of AML (cases), as well as samples from similar individuals who did not develop AML (controls), were sequenced. The researchers found that CH was more frequent in cases than controls. Consistent with earlier studies, more mutations and higher VAFs were associated with higher risk. In addition, mutations in certain genes, including those encoding spliceosome components, TP53 and IDH1/2 displayed a particularly high risk for progression, sometimes even at low VAFs. Remarkably, Abelson et al 40 additionally developed a machine-learning algorithm that used routine complete blood count parameters (eg, leukocyte counts and red blood cell distribution width) from the preceding year to predict risk of AML. Although the data are very promising, screening of the general elderly population (in particular, using large NGS panels covering all the putative driver mutations in AML associated genes) may be premature at this time, as the low overall incidence of AMLs could lead to lower positive predictive values, in comparison to performance in case–control studies.40
CH in other contexts
Small haematopoietic clones observed in situations not directly related to ageing carry different clinical implications, and often different genetic profiles. The present section also includes a discussion of the relationship between CH and CVD.
Molecular MRD and AML relapse
Emerging data indicate that identifying persistent CH after treatment for AML has important implications for patient outcome. As an example of MRD monitoring for the subset of AML with mutated NPM1, Ivey et al 35 developed a reverse transcriptase PCR assay to detect low-level mutations in patients in morphological remission. The authors found that the presence of residual NPM1 mutations was associated with significantly higher risk for relapse and shorter survival.
Jongen-Lavrenic et al 42 studied a broader array of residual mutations in patients with AML in morphological remission via targeted NGS. In this study, while mutations isolated to the so-called ‘DTA’ genes (ie, DNMT3A, TET2 and ASXL1, which are relatively frequent in elderly individuals with CHIP) appeared to confer no obvious adverse outcomes upon follow-up, non-DTA mutations (which are found in biologically more advanced clones, eg, see figure 2) were associated with significantly higher rates of relapse and shorter survival.
Aplastic anaemia
Yoshizato et al 22 identified CH in approximately 50% of AA patients from two large cohorts. The unique genetic profile in AA suggests that clones with specific mutations were selected for in the setting of autoimmunity. On follow-up, clones with mutations in BCOR, BCORL1, and PIGA were more likely to remain stable in size and respond to immunosuppression, and these patients had longer survival. Conversely, clones with mutations in DNMT3A and ASXL1 were more likely to expand over time, and were associated with inferior outcomes.
Therapy-related myeloid neoplasms
Recent research has also modified our understanding of the pathogenesis of t-MNs. While the traditional hypothesis had been that chemotherapy itself causes mutations in haematopoietic stem cells, Wong et al 26 identified very low-level mutations in TP53 (a gene recurrently mutated in t-MNs) using NGS with molecular barcoding in the blood samples of patients prior to the development of t-MNs, and in some cases, prior to the patients receiving any chemotherapy. Interestingly, leukocytes carrying heterozygous TP53 mutations preferentially expanded following exposure to cytotoxic injury in mouse models. Since the overall mutation rates are similar between cases of t-AML and de novo AML, the authors argued that rather than causing mutations, the primary role of chemotherapy in the pathogenesis of t-MNs is to select for pre-existing low-level chemo-resistant clones, including those carrying mutated TP53.
This model is supported by subsequent studies. Takahashi et al 25 and Gillis et al 23 showed that in patients who received chemotherapy, pre-existing low-level mutations, including those involving TP53, were more frequent in patients who eventually developed t-MNs (cases), than in those who did not. In many cases, the small clones were detected using NGS with molecular barcoding prior to chemotherapy. In a similar study, Gibson et al 24 showed that CH was common following chemotherapy for lymphomas given prior to autologous stem cell transplantation (ASCT). The presence of CH in this setting, mostly characterised by mutations in TP53 and PPM1D, was associated with elevated risks for t-MNs and cardiovascular events, and shorter survival, following ASCT. Since mutations may be detectable prior to therapeutic interventions (ie, chemotherapy and ASCT), these could serve as useful predictive biomarkers, enabling clinicians to identify patients at higher risk for developing t-MNs.
The relationship between CH and CVD
In 2014, Jaiswal et al 2 found in their exome sequencing study that CH was associated with higher risks for coronary artery disease and stroke. Since there were very few leukaemia-related deaths in their cohort, the authors proposed that the increase in mortality they observed may be in large part attributable to cardiovascular causes. More recently, Fuster et al 17 and Jaiswal et al 16 both investigated for a causal relationship between CH and CVD, using low-density lipoprotein-deficient (atherosclerosis prone) mice with reconstituted marrows carrying homozygous and heterozygous mutation in TET2. These animals showed expansion of TET2-mutated clones and larger aortic root plaques in comparison to controls on high-fat diets. In Fuster et al,17 TET2- mutated macrophages showed increased interleukin-1 beta production via the NLRP3 inflammasome; interestingly, an NLRP3 inhibitor reduced the size of plaques in TET2-deficient mice. Jaiswal et al 16 similarly demonstrated via expression profiling the overexpression of several chemokines and cytokines in TET2-mutated macrophages, and additionally linked mutations in DNMT3A, TET2, ASXL1 and JAK2 to CVD in four independent cohorts. The overall findings from these studies support the idea that higher cardiovascular risk in CH is caused at least in part by mutant macrophages that drive aberrant inflammation, leading to accelerated atherosclerosis.
Should we test for CH at this time?
Based on existing studies, potential indications for testing for the presence of mutations in haematopoietic cells include the following: risk stratification in patients with cytopenias, patients with cancer who are about to receive chemotherapy with or without autologous stem cell transplantation, prospective bone marrow donors (for the risk of donor-derived leukaemias in recipients),43 patients with AML in morphological remission and individuals at risk for CVD. However, there are many considerations that have not been fully worked out. Importantly, while drugs targeting specific mutations are under investigation,44 it is unclear how best to manage CH in most settings. In other words, there is actually little data at this time concerning potentially beneficial clinical interventions that patients could expect from sequencing their blood samples. From a molecular testing perspective, there is also no consensus regarding how best to test for CH. NGS gene panels will likely require updates as additional biologically important mutations are discovered. In addition, the choice of specific technology platforms will likely vary across laboratories and require consideration of DNA target size, DNA input amount, the expected volume of cases, the numbers of samples that could be run at one time, requirements for analytical sensitivity and variant quantification, and other factors (table 2). For example, higher sensitivity may be preferable for the detection of MRD and small pre-existing TP53 clones in patients who are about to undergo chemotherapy; however, a highly sensitive assay may also detect very small incidental clones in elderly individuals with unexplained cytopenias that may be inconsequential.
Irrespective of this, as the use of sequencing technologies becomes more widespread in clinical diagnosis, a working understanding of CH can be helpful to haematopathologists in practice. For example, sequencing performed on a peripheral blood sample from a patient with cytopenias may reveal CCUS. Testing performed on a bone marrow aspirate from a patient with AML in morphological remission may show mutations that confer a higher risk for relapse. At institutions and laboratories with large pan-cancer NGS panels, somatic mutations in genes associated with myeloid neoplasms (eg, DNMT3A or JAK2) may be detected, incidentally, during sequencing of solid tumours.45–48 Such a result can be difficult to interpret without further work-up. Conceivably, the mutations could have originated from the solid tumours or background infiltrating leukocytes, but without sequencing of a concurrent blood or bone marrow sample, this distinction can be difficult. If the mutation is shown to originate from the leukocytes, a thorough haematological work-up, including possibly a bone marrow biopsy, may be required to separate incidental CH from a concurrent myeloid neoplasm.
Concluding comments
In summary, emerging research suggests that the biological significance of CH depends on the number and identity of mutations, the sizes of mutant clones, as well as, critically, other factors, including patient age, immune status, exogenous drugs/toxins, and perhaps even traditional CVD risk measures. As we learn more about CH, the methods we use to detect mutations will likely evolve in parallel, as clinical needs change. Over time, new methods, ideas, frameworks, and nomenclatures will likely emerge, enabling us to better communicate the molecular underpinnings of myeloid neoplasms.
Take home messages
Clonal haematopoiesis (CH) is defined by the presence of somatic mutations and/or cytogenetic abnormalities in haematopoietic cells.
These clones do not show morphological abnormalities under the microscope, and occur most frequently in elderly individuals with normal blood counts, distinguishing them from myeloid neoplasms in the WHO classification.
CH is associated with the development of haematological malignancies and cardiovascular disease.
The specific clinical implications of CH in different contexts are dependent on the identity and number of mutations, the size of the mutant clones and several other factors (eg, autoimmunity and exposure to chemotherapy).
As we learn more, the nomenclature used to describe CH, as well as the molecular technologies used, will likely evolve.
References
Footnotes
J-MC-C, PM and RT-LM contributed equally.
Handling editor Runjan Chetty.
Contributors JC, PM, RTL and DX contributed to the conception, drafting and revising of the manuscript. SA and RPH provided critical feedback and revised the manuscript. All authors approve the version of the manuscript for submission.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Commissioned; internally peer reviewed.